Molecular Topology
NMRFx Analyst applications are often about studying molecules, so it's not surprising that you can use it to keep track of information about your molecule. NMRFx Analyst supports the use macromolecules like proteins and RNA and the use of small molecules (including ligands of macromolecules). But how do you get all those bonds and atoms into the program where you can do something useful with them. Files containing moleculear structures are generally input through the Molecule->File menu or (for macromolecular sequences) additionally through the Sequence GUI tool.
Reading small molecule files
mol, sdf or mol2 files
NMRFx can read files in these three formats containing one (mol or mol2) or multiple molecules (sdf) files. Just use the corresponding Molecule>File item (Mol for .mol and .sdf, Mol2 for .mol2 files)
SMILES
SMILES is a format that describes the molecular connectivity (atomic elements and the bonds that connect them) in a simple text string. You can enter a molecule in SMILES format using the Molecules->Input SMILES menu item or by reading a file containing SMILES strings.
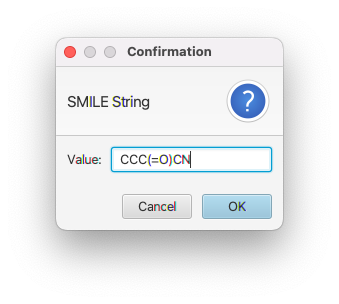
If you use the GUI input you'll be prompted to enter a SMILES string, and then a name for the molecule.
If you read a file containing SMILES strings you can import multiple molecules. Each line of the file should have one or two entries separated by tab characters (if there are two items). If there are two items then the first item is used as a molecule name and the second item is parsed as a SMILES string. If there is only one item, then it is parsed as a SMILES string and a name is automatically generated from the file name and an incremented index for each line.
Reading macromolecular sequences
Sequence File
Macromolecules like proteins, RNA and DNA, are polymers of a small number of standard residues so all you need to do is give NMRFx Analyst a list of the names of the monomers. The simplest format for this information is just a text file containing the names of the monomers. To read the file, use the Molecules -> File -> Read Sequence menu item. This will read the molecule in, and setup up some information within NMRFx Analyst so that when you save a "STAR" file (explicitly or as part of a project) the molecular structure will be saved as well. That way, you only need to explicitly read the molecule in once.
The entries in the file are the three letter names of the amino acids, and one or two letter names for nucleotides. For DNA nucleotides, the name has a first letter of "d" to distinguish it from RNA.
GLY
ALA
MET
ASP
SER
LYS
THREach line can have a residue number as well as the residue name
GLY 1
ALA 2
MET 3
ASP 4
SER 5
LYS 6
THR 7Lines without numbers continue incrementing from the number of the previous residue's number. So this file would have residues with numbers 10, 11, 12,13,16,17,18
GLY 10
ALA
MET
ASP
SER 16
LYS
THR By default the molecule name used within NMRFx Analyst is derived from the name of the sequence file. What if you want your file named myfavoritemoleculein2006.seq, but want to use a simpler name within the program, like Fred? Just put a name for the molecule in your sequence file. To do this you should include a molecule line at the beginning of the sequence file. This line should have two fields, the first should read "-molecule" and the second should be the name of the molecule. Two additional special lines can also be added to further define the molecule. The "polymer" and the "coordset" fields lines have the same format as the molecule line, that is "-polymer" or "-coordset" followed by the name. NMRFx Analyst will use them to allow multiple polymer and coordset entries. Think of the polymer as being a unique amino acid (or nucleic acid) sequence. The coordset corresponds to what X-ray crystallographers refer to as an assymetric unit. A homodimer would have one polymer, and two coordsets.
Ligands can be specified in the ".seq" file with a line like "-sdfile fileName.sdf". The file specified with the name must an ".mol" or ".sdf" file and must be in the same directory as the ".seq" file.
Here are some examples of sequence files
- Simple sequence
- met ala asn glu lys
- Sequence starts at "5"
- met 5 ala asn glu lys
- Sequence with breaks
- met 5 ala asn 8 glu lys thr his 15 arg thr
- RNA sequence
- G
G
C
A
U
C - Sequence with entity names
- -molecule fred -polymer chainA -coordset mono1 met 5 ala asn glu lys
- Heterodimer
- -molecule mymol -polymer poly1 -coordset A met ala asn glu -polymer poly2 -coordset A val asp arg
- Homodimer
- -molecule mymol -polymer poly1 -coordset A -coordset B met ala asn glu
- Polymer with ATP ligand
- -molecule atpBinding -polymer chainA -coordset A -sdfile atp.sdf met ala asn glu lys
- DNA with negative resnum
- -molecule quad da -2 dt dg 1 dc dg
PDB Files
What if you already have a PDB file and don't want to be bothered writing a sequence file. Not too worry, you can read the PDB file directly with the Molecules->File->Read PDB menu item. If you use this command, then NMRFx Analyst figures out what the residue sequence is from the PDB file, and then loads the appropriate residue topologies from NMRFx Analyst's own residue library. The resulting atoms and bonds should be the same as if you had read a sequence file with the same sequence as the that in the PDB file. Because of this, the atom names may not be exactly the same as what's in the PDB file.
If instead, you use the cMolecules->File->Read PDB XYZ menu item, then NMRFx Analyst will use the exact atoms that are in the PDB file. In this case NMRFx Analyst figures out the bonding based on inter-atomic distances and it may not get the bonding exactly right.
If you have a molecular structure loaded already, but want to update the atom coordinates with those in a PDB file then you can use the cMolecules->File->Read Coordinates menu item. NMRFx Analyst will set the coordinates of atoms in the already loaded molecule to be the same as those in the PDB file. Only atoms whose name exactly matches the name of an atom in the file will have their coordinates set.
Sequence Editor
You can also enter a molecular sequence directly within NMRFx without loading a file. Just use the Molecules->Sequence Editor menu item to display a sequence editor. Enter a Type (Protein, RNA, DNA), molecule name, a chain name (defaults to A) then enter a sequence using one letter codes. You can have spaces and line-breaks in the sequence. Then click Add Entity to add a polymer chain with that sequence. You can do this multiple times to add multiple chains to the molecule.
Topology and Coordinates
The Topology from Sequence commands (either reading a sequence file, reading the sequence from a PDB file, or using the Sequence Editor) just generate molecular topology information within NMRFx Analyst. (Whats topology? Just the names of the atoms, and what atoms are bonded to what other atoms.) There is no information about where in space (not to mention time) they are. You can have your topology and coordinates too (if you know what they are). If you've already generated a topology and want to add coordinates use the menu command. The above two methods that use PDB files will get coordinates for each atom they can find in the PDB file
When using the above menu option to read coordinates in NMRFx Analyst will read all the "models" in the PDB file. Each model will be stored in a structure accessible with the specified model number. For example, all the coordinates after "MODEL 4" will be in structure number "4" in NMRFx Analyst. All the models will be used when using the peak identification tools and when calculating atom rmsd values and molecular superpositions. When NMRFx Analyst first reads a sequence file it will automatically generate 3D coordinates for the atoms. These will be for the molecule in an extended conformation and will be stored in structure number "0". Structure number "0" is deselected in coordinates are explicitly read in. To specify which structures are currently active use the Structure Selector described below.