Overview
Processing is set up with controls in the Processor panel at the right side of the spectrum window. This panel appears automatically when you open a raw NMR dataset (FID file) and can also be displayed by clicking the Processor button at top right of the spectrum. This part of the GUI is used for configuring and executing processing scripts.
Load FID
If you display the Processor panel (by clicking the Processor button) and the active chart in that window has a dataset that doesn't have a currently opened FID (that is the dataset was directly opened rather than being generated from processing an open FID), then the panel will contain a single button Load FID. Clicking that button will load the FID that the current dataset was derived from. This only works if the appropriate FID can be identified. For example, NMRFx format datasets processed with recent versions of NMRFx will have a parameter entry that stores the path to the FID. But older ones, or files processed in other applications, won't. If there is no parameter containing the FID file location a file browser will appear to select an FID file.
Simulate FID
If you display the Processor panel (by clicking the Processor button) and the active chart in that window doesn't have a dataset, then the processor will contain a tab for use in generating simulated data vectors. This is useful for educational purposes, and allows students to simulate and process FIDs.
Process FID
If you display the Processor panel with a FID present, or if it is displayed when you open an FID dataset, then you'll get the full tool with multiple titled panes and controls.
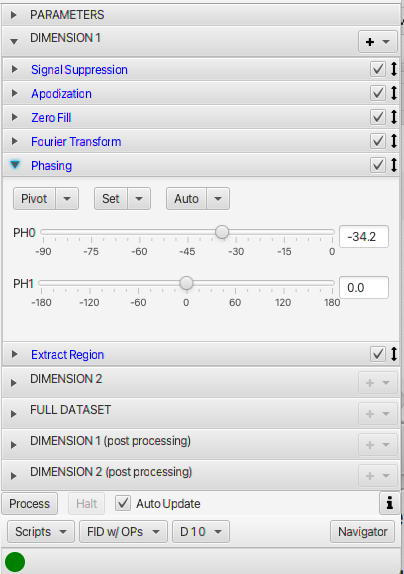
The Processor panel has a series of titled panes and a set of controls at the bottom. The titled panes open when you click on the title bar. The top titled pane (labelled PARAMETERS) allows you to view and change (some) dataset parameters. These include solvent, pulse sequence, temperature and dimension specific values like sizes, spectrometer frequency and sweep width. The use of the parameters is described on the Parameter Pane page. The next series of titled panes (labeled DIMENSION 1 etc.) contain a series of operations that carry out the actual processing (apodization, Fourier transform, phasing etc.). The use of Dimension Processing panes are described on the Operations page. The controls at the bottom of the panel are used to trigger processing, interact with scripts or change the viewing mode. The use of the controls at the bottom of the Processor are described here.
- Process
- Click this button to apply the current processing operations to the current FID.
- Auto Update
- If this is turned on (the default for 1D and 2D datasets), then whenever a processing operation is added, removed or its parameters changed the current processing operations will be applied. If it is turned off, then use the Process button to trigger processing. The Auto Update allows rapid feedback of the effect of the operation changes, but is only feasible if the dataset is small enough that processing can be completed rapidly (ideally 1sec or less).
- Halt
- Click this button to halt long running processing. It is only active while processing is active.
During processing an indicator at the bottom of the window will display the progress of processing. If processing is completed successfully the elapsed time will be displayed at the bottom of the window (next to status circle). If an error occurred the status circle will be red, and an error message will be displayed next to it. If you click on the red status circle a window will open to display error information.
Controls at the top of the processor can be used to change the active dimension and whether or not the processed multidemensional dataset (.nv file) is displayed.
- View Mode
Switches between viewing the FID or processed dataset
FID
The display is the raw FID, without any operations applied
FID w/OPs
The display is the FID, with any active operations applied. You can turn on and off operations to see there effect on the displayed FID.
Spectrum
- The fully processed data (in all dimensions). This represents the processed data as output to the output file.
In FID, and FID w/OPs modes the displayed FID depends on what dimension titled pane is currently open. For example, with Dimension 1 opened, the direct (first) dimension of the dataset will be shown. If the dataset has more than one dimension and the Dimension 2 panel is opened then the first indirect (second dataset dimension) will be shown. NMRFx Analyst is capable of extracting a vector along indirect dimensions so that one can see and observe the effect of processing operations not only on the directly detected FID, but also on indirectly detected FIDs. The utility (and sometimes capability) of showing these "indirectg FIDs" will depend on the type of experiment and its sensitivity. But for many experiments it can be useful to do initial set up of the phases of the indirect dimension.